Hidden inside our DNA are many short sequences that repeat again and again, like stutters in the genetic code. Most of the time, these repeats are harmless. But when they grow too long, they can disrupt how genes work and trigger serious neurological and developmental diseases. These conditions, known as repeat expansion disorders, are now at the center of a quiet revolution in genomic medicine.
Some of the most well-known genetic diseases belong to this group. Huntington’s disease is caused by an expanded “CAG” repeat in a single gene. Fragile X syndrome, the most common inherited cause of intellectual disability, arises from an expanded “CGG” repeat. Spinocerebellar ataxias, myotonic dystrophy, and several forms of early-onset dementia all share the same basic problem: DNA repeats that have grown far beyond their normal length.
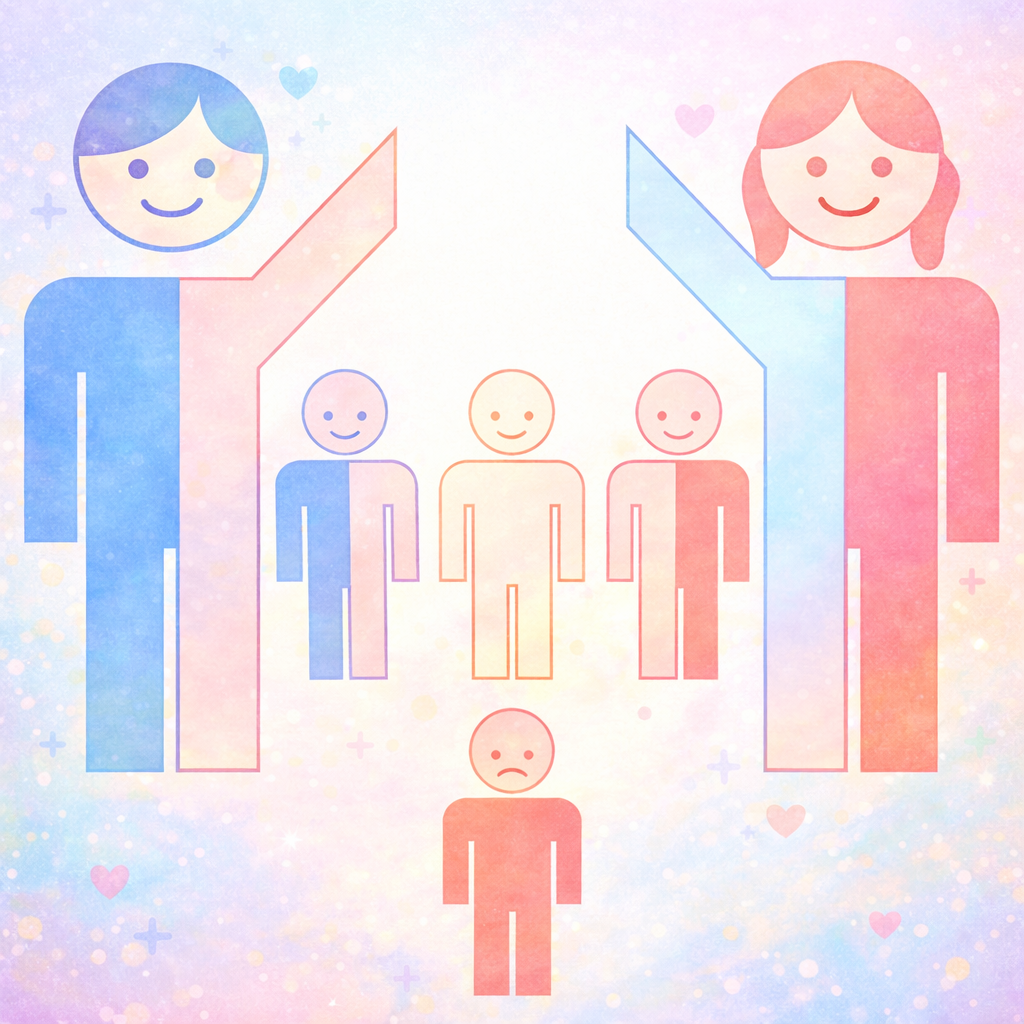
What makes these diseases especially striking is that they often worsen across generations. A parent with a mild form may have a child with earlier onset or more severe symptoms, because the repeat tends to lengthen when passed on. This phenomenon, called anticipation, explains why some families see neurological disease appearing younger and stronger with each generation.
For decades, diagnosing these disorders required special, gene-by-gene tests. Each suspected disease needed its own assay, and only a handful of known repeats could be checked at a time. Even then, many patients with unexplained tremors, memory loss, psychiatric changes, or coordination problems remained undiagnosed. The repeats were simply too long, too complex, or located in regions that standard sequencing could not read accurately.
Whole-genome sequencing (WGS) is now changing this story.
Modern WGS technologies can scan the entire genome and detect repeat expansions across dozens of genes in a single test. New analytic methods can estimate repeat length, identify abnormal patterns, and flag expansions that were previously invisible. This means doctors no longer have to guess which disorder to test for first. One genome sequence can screen for Huntington’s, fragile X, multiple ataxias, and many newly discovered repeat diseases at the same time.
This has led to important new discoveries. In recent years, scientists have linked repeat expansions to forms of amyotrophic lateral sclerosis (ALS), frontotemporal dementia, epilepsy, and even some psychiatric syndromes. Many patients once labeled with “idiopathic” neurological disease are now receiving precise genetic diagnoses because WGS finally reveals the hidden repeats.
The impact goes beyond diagnosis. Knowing the exact repeat length helps predict disease severity, age of onset, and risk to family members. It guides genetic counseling, reproductive planning, and, increasingly, clinical trials aimed at silencing or shrinking toxic repeats.
Perhaps most exciting is what this means for the future. As WGS becomes routine, repeat expansions may be detected earlier, sometimes before symptoms begin, opening the door to monitoring and early intervention. Therapies targeting RNA toxicity and abnormal proteins are already moving through clinical trials.
Repeat expansion diseases remind us that genetic errors are not always simple mutations. Sometimes, disease comes from the same word written too many times in our DNA. With whole-genome sequencing, medicine can finally read those repetitions clearly, and, at last, begin to break their spell.